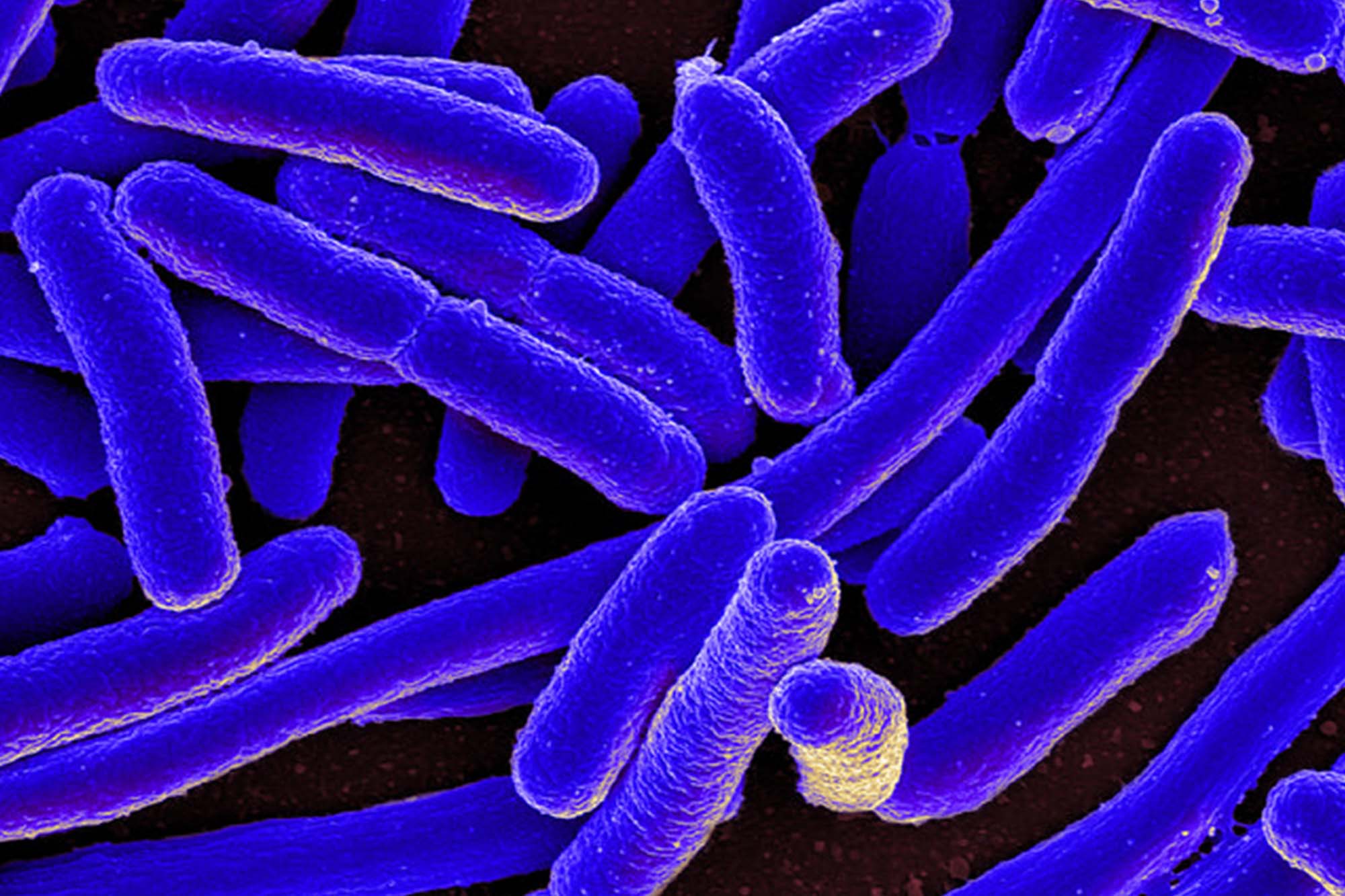
Motile bacteria are capable of swimming efficiently toward favorable chemical environments and away from inhospitable ones. This behavior–called “chemotaxis”–is frequently used by unicellular organisms for finding food.
Not surprisingly, such behaviors play important roles in establishing beneficial host symbioses and pathogenic infections. The value of understanding in detail this mechanism of directed cell migration in response to extracellular chemical signals cannot be over-stated, and Escherichia coli, commonly referred to as E. coli, has become the paradigm molecular model.
“Despite their tiny size, bacteria have evolved amazingly sophisticated protein machines for detecting and responding to changes in their environment,” says John “Sandy” Parkinson, principal investigator in the School of Biological Sciences. Caralyn Flack, a research associate in the Parkinson Lab, developed a technique for monitoring the behavior of living bacteria before and after she changed the structure of a critical signaling protein inside the cell.
Flack’s new findings were recently published in Proceedings of the National Academy of Sciences (PNAS), a peer reviewed journal of the National Academy of Sciences (NAS). The paper, titled “Structural signatures of Escherichia coli chemoreceptor signaling states revealed by cellular crosslinking,” makes an important advance in elucidating the molecular mechanism(s) of signal propagation through chemoreceptor molecules.
“Bacteria are amazing at sensing and responding quickly to changes in their environment,” states Flack. “This makes them a great model for trying to understand transmembrane signaling.” But it turns out that changes that accompany signaling events in chemoreceptors are difficult to follow with traditional structural methods, and those techniques that do work cannot replicate the native cellular environment. To remedy that, the technique Flack developed does three things. First, it assesses behavior. Second, it changes the structure of a protein inside living bacteria, and third, it then watches the signaling behavior change in real-time. The technique, which involves “crosslinking,” she says, has proven to be “really powerful for structure-function analyses of signaling proteins.”

In essence, what Flack did was create receptor proteins with a special amino acid at a position of interest. That amino acid, cysteine, has unique chemical properties that, under the right conditions, allow it to form a covalent bond (to “crosslink”) to a nearby cysteine in the same protein or in another protein. Such crosslinks constrain the structural movements of the proteins and can change its behavior. This is how Flack was able to change the structure, and in consequence, the function of a receptor within a living cell.
The crosslinking reporter sites “enabled us to evaluate receptor output states before and after crosslink formation,” she writes. This new cellular crosslinking approach “delivers unprecedented insight into receptor structural properties as the receptors function within cells” stated one reviewer of the PNAS manuscript. Flack suggests that “similar crosslinking approaches could serve to follow signal transmission in other regions of the chemoreceptor molecule and perhaps in other signaling proteins, as well.”
Work on those topics is ongoing.
You can read more about Caralyn Flack and her work in the Parkinson lab here.
