
Last semester, Professor Colin Dale’s Biol 5275, Microbial Diversity, Genomics and Evolution (MDGE) lab/class was faced with an especially ambitious task.
Students were challenged to isolate culture caffeine-degrading bacteria from environmental samples, perform molecular identification, sequence their genomes and identify the genes responsible for degradation of caffeine. They explored a diverse and interesting range of starting materials, including coffee maker drip trays, soils, live caffeinated plants and coffee beans from across the globe. They also obtained a couple of SCOBYs (symbiotic culture of bacteria and yeast), an ingredient used in the fermentation and production of kombucha tea.

“These upper-class students have been tremendous this year,” Dale says, “with an excellent cohort who all completed what many might consider to be an overly-optimistic set of goals for a semester.” Indeed, the pandemic has driven the class in a more independent direction. Since it’s not been possible to travel together to a field site, such as the Great Salt Lake or a local hot spring, projects have been devised that allow the students to collect environmental samples on their own. This involves the instructor, Dale, describing a basic goal (to collect and characterize bacteria that can degrade caffeine) and the students developing a scientific hypothesis to address the goal. After 10 weeks of laboratory work, the students prepare and present a poster, which means, in just one semester, they devise, conduct and present a whole scientific project!

The research was presented in the traditional poster format on December 8, 2021 in the lobby of the Aline Skaggs Biology Building where the SBS community was invited to attend, ask questions, and witness the results that, in some cases, challenged the very dogma of traditional microbiology. Not bad for a group of undergraduates faced with a pandemic taking the precautions necessary to prevent transmission in a classroom and lab setting.
One of those COVID-generation students is Alex Smith, a senior, who with his research partner Jayden Sorenson, found some striking results in their work which started out with the plating of samples from a tea plant, a Dwarf Yaupon Holly plant, a coffee plant and guarana. A control plant containing no caffeine was also used.

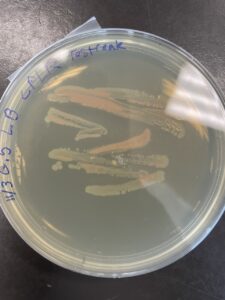
After culturing caffeine-degrading bacteria on agar plates, the young scientists used PCR and other methods to determine the identities of the bacteria prior to full genome sequencing. If a PCR was successful, a clean up process was performed and the resulting DNA was submitted for 16S Ribosomal RNA sequencing. From there a single colony was selected for genomic DNA isolation. Identifications were documented and it was then that a red pigmented colony was observed from the coffee plant leaf which was confirmed as a bacterium.
“A weird finding,” Smith says, “was when the sequencing results came back from this red colony the sequencing failed after multiple attempts. I wasn’t really sure, why.” That was when the scientific hunch emerged, that commonplace but ineffable vector that scientists find themselves subject to when they are stumped. Smith needed more data—or in this case more microbial growth—so he streaked the red pigmented colony onto another plate in an attempt to make it genetically clonal and ensure it fed on caffeine. Following that, it was subsequently streaked onto a nutrient-rich LB plate. This resulted in significant growth but now there were two differently pigmented colonies visible on the plate!

Professor Dale was curious to see the genome sequencing results. When they were obtained and analyzed it became clear that there were two bacterial genomes — likely arising from the red and yellow colonies. “There were two different genera of organisms that belong to two different families,” says Smith–Cellulomonas and Methylobacteriaceae. Once the team had ruled out contamination of the LB plate they determined, under the direction of Dale, that the two bacteria were each benefitting from the other, a form of mutualism called a “syntrophy.”
Of the two the red Methylobacteriaeceae was the caffeine degrader and the yellow Cellulomonas was clearly not only benefitting from the red but giving back in some supportive way to its red partner, likely providing “some auxotrophic provision that is necessary for growth on a minimal medium,” according to text from the poster. The two organisms clearly needed each other to function.

Whether this is a full-fledged discovery worthy of publication (after additional research and testing, of course) is unclear. But its discovery in the sandbox of the Colin Dale lab reiterates why scientists do what they do… and how.
This is the 17th year that Dale has taught MDGE, which examines the role of microorganisms and their complex interactions with other living organisms and the environment. The lecture course provides an integrated vision of genome biology and microbial physiology, diversity and ecology and serves as a primer for students interested in genomics. In addition, the synergistic laboratory class, the results of which were the subject of the poster presentation, provides students with an opportunity to collect samples from the environment and examines microbial diversity using modern molecular biological methods and bioinformatic tools.
To accommodate pandemic constraints, poster presenters were divided into three 40-minute sessions to keep crowding to a minimum. Even so, spirited discussions ensued, even if presenters, mostly in pairs, had to be asked to speak up from behind their masks. It’s a sign of the times that budding scientists at the School of Biological Sciences must factor the vagaries of a viral pandemic and its emerging variants into their education and research. It’s a tribute to their tenacity and creativity that they were able not only to engage in a research project but to present it so crisply and authoritatively.
As for Alex Smith, the Sandy, Utah native also works as a lab assistant for Professor Dale outside of the 5275 class/lab, the completion of which will propel him towards graduation in the Spring. The experience in the lab with graduate student researchers has sparked an interest in Smith in furthering his own education as a graduate student himself. If not grad school, after graduation Smith may parlay his experience at the UofU into forensic science, but will pursue it through the lens of and expertise for the life sciences.
Clearly, he’s been bitten by the bug . . . or, more accurately, “bitten” by bacteria.
By David Pace
For more photos of the event click here.
Undergraduate biology students like those in Biol 5275 have access early on as freshmen to get hands-on research experience through the Science Research Initiative. You can donate to this scholarship fund here.
