
A RETROSPECTIVE VIEW
by Baldomero Olivera
I have no formal academic training in pharmacology. It’s no surprise, therefore, that my path to a research career in this field has been idiosyncratic. Ultimately, my increasing involvement with pharmacological science came about because it was the indispensable scientific discipline required. At critical moments in my scientific life, recurring themes of cultural difference, technology (or lack thereof), and a kind of naiveté took me down a particular path that, although quite conflated at times, led to pharmacology. These features of my career, along with a personal narrative conveniently framed as a series of serendipitous events, brought me to the distinctive flavor of pharmacological science that has become the guiding principle for my scientific work.
The Advantages of Being Naive
An important factor in my research career is that I have launched projects not as an expert but as a novice. Due in part to my cultural background, I have been willing to enter new fields without much expertise. I am not uncomfortable starting out as an outsider in a new field of science, having grown up under the cultural differences of two educational systems. The Philippines, where I was born, had a 300-year colonial history of Spanish authoritarian rule. This left a highly hierarchical culture in which the social norm was to be deferential to authority. However, I was heavily influenced by the American education system and acculturated to American ways. Although it can be stressful and disorienting to be a foreign-born graduate student in the United States, as I was, my multicultural perspective provided a more flexible and sometimes more unconventional approach to solving research problems.
My graduate work had been in biophysical chemistry, and when I came to the Department of Biochemistry at Stanford University as a postdoctoral fellow in 1966, one advantage I had was that, in addition to focusing on the enzymology that I was attempting to do, I was able to think about the substrate, DNA. A major lesson I had learned from my studies of the biophysical nature of DNA in solution was that single- and double-stranded DNA had different properties, and I believe that this insight worked to my great advantage when setting up the assay that ultimately led to the discovery of Escherichia coli DNA ligase, because the enzyme requires a double-stranded DNA substrate.
A similar novice approach turned out to be an advantage when I decided to study the toxins that are present in Conus venoms. I hadn’t realized that almost all laboratories studying animal venoms characterized whole venom. Not knowing that this was the norm, and given my newly gained biochemical background as a postdoc, I had assumed that the first order of business was to purify the individual components. The naiveté of “purification first, characterization later” was the key to the much more rapid progress we made on cone snail venoms; it is difficult to gain basic physiological insights without having the individual components well defined. Retrospectively, this seems obvious, but this was not how the field of toxinology operated. As a novice in a new scientific field, I had to overcome difficulties. I still remember the scathing grant review when I first tried to submit the work on cone snail venoms for funding. Essentially the review said that I had no expertise in toxinology, no expertise in peptide chemistry, and no expertise in neuroscience and that I was singularly unqualified for the proposed work. This was all true of course, but in the long term my lack of expertise turned out to be an advantage because I was not constrained by the dogma in the field.
Opportunities for Scientific Exploration
As I arrived at Stanford for my postdoctoral studies, a recent development in biochemistry was the characterization of an enzyme, polynucleotide kinase, which made it possible for researchers to label DNA strands at the 5′-end with radioactive phosphorus-32 and to detect any changes that occurred at the 5′-end of a DNA strand. My advisor, Robert (Bob) Lehman, had reasoned that anything that happened to the 5′-end would be of great interest (except if the 5′-end label were degraded). Having a label at that critical position in DNA was the technical advance that opened the door to open-ended exploration; we had no way to predict what might be found.
In the first experiments I carried out, the 5′-end was quickly degraded, released either as phosphate or as small nucleotides. I tried a variety of different types of DNA labeled at the 5′-phosphate and got nothing but degraded DNA. Finally, I decided to try double-stranded DNA with a long strand that had only adenine bases (poly dA) and short strands that had nothing but thymine bases (poly dT). I could easily label the 5′-end of all the short strands. When I made this double-stranded DNA substrate, I found that the 5′-end of the short strands was rapidly transformed after a short incubation. What had happened was that all the short strands were joined together by a then-novel, uncharacterized enzyme, E. coli DNA ligase. This finding ultimately led to the purification of DNA ligase and the elucidation of the mechanism of DNA ligation (1–3). The availability of DNA ligases was one important technology for the recombinant DNA revolution.
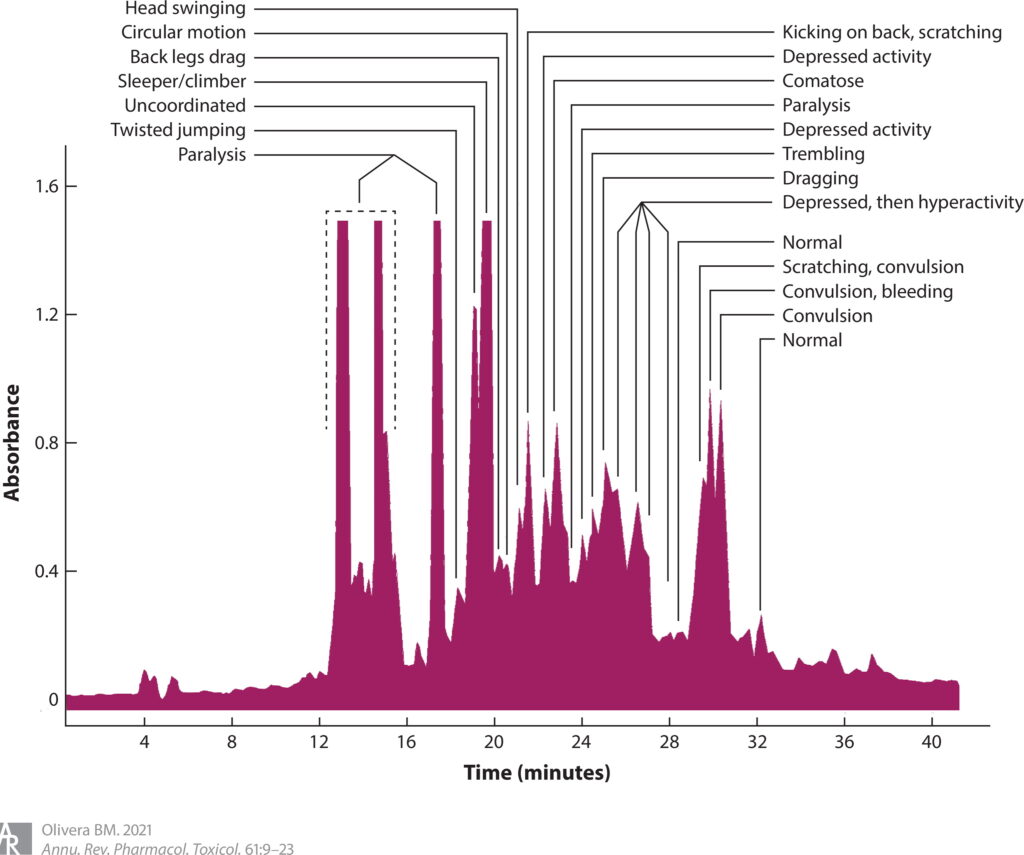
Similarly, when I started to work on cone snails (see Figure 1) and their venoms, a seemingly minor technical advance triggered a period of open-ended exploration where we could not predict what we might find. The key development was low tech—an undergraduate student at the University of Utah, Craig Clark, showed that if he took components of a cone snail venom, he could elicit dramatic changes in the behavior of a mouse (4), provided that the injection was administered directly into the central nervous system. (Craig had learned to inject young mice by the intracranial route.) Up to that point, we had injected mice with venom fractions only through the intraperitoneal route and observed only paralysis or occasionally mild hyperactivity. However, via the intracranial route, the entire horizon was dramatically changed; each of the different fractions of a cone snail venom elicited a different behavioral effect. This made it possible to purify the component that caused a particular change in mouse behavior; for example, there were venom components that made mice go to sleep, others caused scratching, some made the mice run around in circles, and others caused hyperactivity (see Figure 2). Thus, a simple technical advance allowed us to broadly explore cone snail venoms. It was this technical breakthrough that eventually led to the purification of a class of venom peptides called ω-conopeptides (5), one of which ultimately became Prialt (ziconotide), a drug approved to treat intractable pain (6). Prialt is probably the only commercial drug for pain discovered and characterized by undergraduate students.

Thus, my research career has seen a series of open-ended exploration in new areas, with totally unexpected results. This is obviously not the typical paradigm for hypothesis-driven science. In essence, my research trajectory has been determined by expeditions into discovery.
Lucky Breaks at Key Moments
My family was a professional middle-class family with no scientific tradition at all. A first lucky break was made possible by my dad’s decision to take a job in San Francisco, California, in 1947. My dad, Baldomero T. Olivera, was a newspaperman, and because of the massive destruction caused by the Second World War in the Philippines, opportunities were limited, which made the position of press attaché in the Philippine Consulate attractive. Taking this position meant that we had to move to California for one year, and so I was enrolled in Grattan Elementary School, where I met Miss Uhler, my second-grade teacher. She taught all subjects, including science. In one science class, she gave us several powders and asked us to do an experiment: Shake each powder with water. Two of the powders (sugar and salt) dissolved easily in water, but one powder was calcium carbonate, which did not dissolve at all. Other powders gave a colloidal suspension. I still remember that this experiment was a revelation—there were properties of things around you that could be revealed by doing an experiment. Back in the Philippines, my third-grade English class was assigned to write a theme for the school yearbook on what we wanted to be when we grew up. I wrote down that I wanted to be a scientist and described the experiment that I had done in the second grade; I never wanted to be anything else but a scientist.
In my junior year of high school, I transferred to a new experimental university high school. The new school had recruited teachers who wanted to develop something different from the regular curriculum, and we were the first class in the new school. Our science teacher was Dolly Hernandez. She developed both a chemistry class and a zoology class, which became by far my favorite classes. Dolly Hernandez (who later obtained a PhD in Science Education) was a restrained, always proper, but charismatic educator. One day, she brought in a TIME magazine article with Lee A. DuBridge, the president of the California Institute of Technology (Caltech), on the cover. She gave me the magazine, told me to read the article, and simply said, “You should go there after you are done with college.” It was because of Dolly Hernandez that I decided to major in chemistry as an undergraduate at the University of the Philippines. And it was because of Dolly Hernandez and my complete trust in her advice (and also my cluelessness and naiveté) that as I was mulling over what to do after college, when the opportunity came to apply for a fellowship to do graduate work in the United States, I applied to only one graduate school, Caltech. Luckily, they accepted me.
At Caltech, I had to choose an advisor within one month of my arrival in September 1961; most incoming Caltech chemistry graduate students had gotten advice from their mentors as undergraduates, but I had no idea about which lab I should join. I was advised to make an appointment with each professor with whom I might be interested in working in order to help me decide. For most of the interviews, the science described was above my head. However, as an undergraduate I had done a paper on DNA; I found out that Norman Davidson had recently switched from doing kinetics in shock tubes to studying the biophysical properties of DNA. After I interviewed with him, I knew I wanted to work on the projects that he described, but I hesitated. The other entering graduate students had told me that Norman was recovering from cancer; he had been the chemist who had purified uranium-238 in the Manhattan Project, and at the time, no one realized the dangers of exposure to high amounts of radiation, so everyone on the team had come down with cancer. The other graduate students warned me how potentially damaging it would be to my graduate career if my advisor grew gravely ill and I had to switch labs. After a long period of indecision, I had the gut feeling that there wasn’t any other project that I really wanted to work on, so I signed on. That was certainly the most critical decision I made in my scientific life. My thesis work was on the biophysical properties of DNA in an electric field. I had no idea at the time what a singularly exceptional place Norman’s lab was to train for a research career in biology at the molecular level (the field of molecular biology was not yet a coherent and well-defined discipline). Not only did I benefit from Norman’s beneficent mentorship, but the lab attracted the types of graduate and postdoctoral students who created an atmosphere that was both intellectually rigorous and wildly creative. (James C. Wang, who later discovered DNA topoisomerases, and Ron W. Davis, whose work laid many of the foundations for genetic engineering, were some of my contemporaries in the Davidson lab.)
Just as my confidence in the advice of Dolly Hernandez led to the graduate program of Caltech, my confidence in the advice of Norman Davidson led me to a postdoc at Stanford in 1968. Norman felt that the biophysical chemistry I had done would probably be impractical to pursue, because it was my intention to return to the Philippines, and that enzymology would be more feasible. He recommended that I go to Stanford and work with Bob Lehman; I heeded this advice without hesitation.
Good fortune smiled on my postdoctoral studies at Stanford in several ways. First, working with Bob Lehman was a remarkable experience in the context of the tightly knit Stanford biochemistry department. The department was collegial and organized so that everything was shared (this was the vision of the founder of the department, Arthur Kornberg), but it was also extremely competitive and a somewhat scientifically aggressive environment. Working with Bob was a haven from the more tumultuous environment outside his lab; Bob’s lab gave the rich benefits of all the exciting work being done across the department while providing a uniquely protective, relaxed, and stress-free atmosphere. In many ways this was because of Bob Lehman’s previous experience in life—he had come from a modest immigrant background and was drafted as an infantryman in World War II as soon as he came of age and ended up being seriously wounded in combat. As he lay in the hospital, he received orders to return to the front; he did not think he would survive the year. But he did survive, and being an army veteran gave him the opportunity to go to college and ultimately to Arthur Kornberg’s lab. Bob was the first author of the classic paper on the characterization of DNA polymerase (7).
The juxtaposition of almost being killed in the war and having a research career that he loved made him infinitely tolerant, so that even when there was a crisis in the department, Bob, always mild-mannered and positive, reassured everyone that things would turn out okay. I think his wartime experience colored his attitude so that any problem he encountered after that seemed trivial by comparison. He carried that basic cheerful optimism into experimental science. This optimism has influenced my work: No matter how bad things look after an experiment, something positive will come out of it. As a postdoc, I was able to do the first characterization of E. coli DNA ligase, to determine that it used nicotinamide adenine dinucleotide (NAD) as an energy source, and to identify the covalent intermediates in the DNA ligation reaction (3). It was, by any standard, a most successful time, and I probably could have had my choice of the plum faculty positions that were then available at all the major universities. However, I had come to the United States with the intention of returning to the Philippines, so I accepted a job at the Department of Biochemistry at the University of the Philippines, College of Medicine.
Then came a completely unexpected series of events. I got a call from K. Gordon Lark, then at Kansas State University, inviting me to give a seminar. He had called Bob Lehman and told him he wanted to offer me a faculty position; Bob told him that I had a position at the University of the Philippines. The result was that Lark invited me for a seminar, aware I had accepted a position, but laid out a plan in which I could assume my full-time position in the Philippines and, for 4 months of the year, come to Kansas State University and establish my own independent lab. The arrangement he proposed had come about in a curious way. As an incoming college freshman, Gordon Lark was assigned an upper classman who would orient him when he arrived at the University of Chicago. Every new student had such a mentor; it turned out that Gordon’s was James Watson. Gordon and Watson remained friends and overlapped at the Cold Spring Harbor laboratory every summer. When Gordon wanted to recruit a collaborator to Kansas State University, he had called Watson for suggestions.
A few months prior to that fateful phone call, I had gone to the annual Cold Spring Harbor Symposium in 1966. I had just finished my characterization of the DNA ligation reaction. It was an exciting time for me; I got to meet Fritz Lipmann, who had clarified the role of ATP as the energy currency of the cell. He was interested in the discovery that DNA ligase uses NAD, not ATP, as a cofactor. James Watson was at Cold Spring Harbor, and he was sitting in the front row. Much to my dismay, when I gave my talk, I didn’t see his face at all. He was reading a newspaper the whole time! Apparently, this was typical of Watson during scientific talks, but it didn’t mean that he wasn’t paying attention. I guess he must have been listening, because when Gordon called, Watson strongly recommended me, which is why Gordon decided to offer me this unprecedented part-time faculty position. This unusual arrangement was characteristic of Gordon; he had an unparalleled ability to pursue unconventional solutions when seemingly insoluble problems arose. From 1969 to 1972, I spent most of the year in the Philippines and moved annually with my wife, Lulu, and our two children, Felicia and Jon, first to Manhattan, Kansas, and later to Salt Lake City, Utah, when Gordon accepted the position of chair of the newly created Department of Biology at the University of Utah. The university has now become a flourishing center of biological and biomedical research. The nascent research community, in fields such as molecular biology and community ecology, was built by Gordon Lark in creative, often wildly unconventional small steps; he was ultimately able to hire 17 junior faculty members, and he personally mentored all of us all carefully. It was his guidance that makes me feel unconstrained when exploring unusual solutions to seemingly intractable problems.
Encounters with Cone Snails
Starting my independent research by running two different laboratories was the major factor that led me to a pharmacologically oriented career. The lab in Kansas continued the work in the field of DNA metabolism and focused on how discontinuous DNA synthesis was carried out as well as on nonoxidation/reduction reactions that involved NAD (8–15). I had wanted to carry out the same work in the Philippines, but this was not possible. In contrast to my lab at Stanford, where I had access to any equipment I might want (because the Department of Biochemistry at Stanford was arguably the best-equipped biochemistry department in the world at the time), I had an empty lab with no equipment. I had to find a research project that I could start in the absence of any equipment, which was what led me to study cone snails.
This decision was biased by events in my childhood. After returning from the United States, my parents decided to buy a home in a suburb of Manila that was completely undeveloped—we were the only home at the end of a long road, without neighbors close by. I didn’t have any siblings, and there were no other kids around. Because we lived in an isolated home and had no access to public transportation, I would meet my dad at the tennis court near my elementary school. He often had not finished his match when I arrived. At the time, tennis courts in the Philippines were coated with crushed shells dredged from Manila Bay—before the material was crushed, it lay in piles close to the court. I had nothing to do while I waited for the tennis game to end, so I would sort through the piles and often find shells with interesting patterns and unusual colors, which I started to bring home. As a result, shell collecting became my hobby. My interest in shell collecting grew more focused, particularly after I had Dolly Hernandez as my zoology teacher; she infused my nascent interest with significant scientific content. Even in high school, I had read books and articles on the animals that made the shells, and I knew that some of the cone shells in my growing collection were made by venomous snails, some of which could kill people.
While I was considering what to work on in my empty Philippine laboratory, I decided on the venoms of cone snails. I thought that by using a biochemical approach, we could explain why a snail venom might kill people. I fully expected this to be a short, stop-gap research project that I would carry out only until I could get the equipment required for studying DNA.
CONE SNAILS AND THEIR VENOMS: FROM BIOLOGY TO BIOMEDICAL APPLICATIONS

The decision to start research on cone snails conferred a hidden advantage: I had the perspective of two different scientific cultures. All of my training, as an undergraduate in chemistry, as a biophysical chemist in graduate school, and as an enzymologist, was at the molecular level, with a reductionist perspective. But after I picked up my first cone shell in the pile of dredged shells near my dad’s tennis court, I had become increasingly absorbed by the cone snails. My desire to learn everything I could about cone snails, from their taxonomy and phylogeny to their evolutionary history, was insatiable, and I was familiar with what little knowledge there was about their ecology and behavior. This duality of perspectives gave me a constant stream of facts to mull over and try to connect, and it has been most gratifying for me to take some new molecular detail revealed by the latest lab results and link this to the wider biology of cone snails, or to do the reverse—to learn something new about the biology of a particular cone snail species and to rationalize why that might explain the presence of specific molecules I knew had evolved in their venoms.
Overview of Cone Snails and Their Venoms
There are approximately 1,000 living species of cone snails, and all of them are venomous predators. Each cone snail specializes on specific prey, with venom as the main offensive weaponry. (Cone snails also use their venoms for defense against predators and to gain an advantage over competitors.) The venom of cone snails is complex, with a repertoire of several hundred different bioactive components. These are mostly small peptides, ranging in size from 10 to 40 amino acids, most of which are highly structured by multiple disulfide cross-links. What also became quickly obvious was that every cone snail species has its own distinct complement of bioactive peptides, with essentially no molecular overlap between the venoms of two different species. Thus, the genes that encode these small peptides are subject to accelerated evolution, with the entire complement of hundreds of genes expressed in the cone snail’s venom gland evolving so rapidly that the entire set of gene products becomes biochemically distinct as speciation occurs. This rapid divergence at the molecular level reflects the divergent biology of different cone snail species.
In the Philippine lab, we initially analyzed two fish-hunting cone snail species (approximately 10–15% of all species hunt fish; the others prey on worms or other snails). The first species was the geography cone, Conus geographus. As a shell collector, I had known that this snail could kill people; therefore, it was an attractive starting point for our research, because the geography cone snail venom was bound to have highly potent components that would be easy to detect. The second species analyzed in parallel was the magician’s cone, C. magus. This smaller, extremely common species is routinely sold in markets for food in the central Philippines. People who have tasted both species say that C. magus tastes good in a coconut soup with ginger, garlic, and other spices, but that C. geographus has a somewhat bitter aftertaste. Both cone snails are common in the Philippines, so it was easy to accumulate enough venom glands for biochemical work.
As the biochemical work was being initiated, we maintained a few specimens of both C. geographus and C. magus in our aquaria—visitors to the lab were fascinated to watch the snails hunt fish. C. geographus and C. magus had different strategies for capturing their fish prey. C. magus would extend its long proboscis toward the fish and try to sting the fish and inject venom using a harpoon-like tooth. When the tip of the proboscis touched skin, the snail would jut out the tooth, piercing the fish skin. Powerful circular muscles at the end of the proboscis grasped the end of the tooth, effectively tethering the fish (Supplemental Video 1). However, the tooth is both tether and hypodermic needle. Once the fish skin is pierced, the snail begins injecting venom. The effect is almost immediate—the fish is immobilized and its fins stiffen, allowing the snail to draw the fish toward its false mouth, where the paralyzed fish is digested (16). Several hours later, the snail regurgitates the scales, fish bones, and the harpoon-like tooth used to tether prey. C. geographus uses a different strategy (17). As soon as a fish is introduced into a tank, the snail becomes extremely active, and as it approaches a fish, it opens up its false mouth in the direction of the fish; it apparently releases venom components into the water, and as a consequence, the fish does not swim away, allowing the snail to completely engulf it (Supplemental Video 2). Venom is injected into the engulfed fish, causing an irreversible paralysis.
As described in the next section, both snail species have a cocktail of paralytic toxins that act on voltage-gated calcium channels, voltage-gated sodium channels, and the nicotinic acetylcholine receptor (nAChR). These toxins cause the paralysis of the fish. In the case of C. magus, these are not the same toxins that cause rapid immobilization. Rather, another set of toxins that target a different set of voltage-gated ion channels (Supplemental Video 3) trigger a massive depolarization of all the nerves present close to the injection site of the venom (16). The effect is similar to that of a taser; the fish is effectively stunned and immobilized, allowing the snail to reel it in toward its mouth by retracting its proboscis.
Characterizing Bioactive Venom Components
All the initial biochemistry was done with Lourdes J. (Luly) Cruz, another faculty member of the Department of Biochemistry at the University of the Philippines, College of Medicine; she had done PhD research at the University of Iowa and postdoctoral work at the International Rice Research Institute in the Philippines. Initially, we assayed for paralytic activity on mice and purified several active fractions from C. geographus venom (4, 18); these were α-conotoxin GI, a 13–amino acid peptide (19), and μ-conotoxin GIIIA, which has 22 amino acids (20). We collaborated with several electrophysiologists, notably John N. Barrett at the University of Miami and Doju Yoshikami at the University of Utah, to show that α-conotoxin GI and μ-conotoxin GIIIA inhibited nAChRs and voltage-gated sodium channels, respectively. These were the first bioactive Conus peptides characterized.
As I noted above, the big breakthrough in our biochemical work was the assay developed by Craig Clark—when we analyzed C. geographus venom, we discovered that fractions that were not paralytic elicited striking behavioral changes in the mice upon intracranial injection. Notably, the sleeper peptide elicited a sleep-like state, the shaker peptide caused a characteristic tremor lasting many hours after injection, and the sluggish peptide made mice hypoactive. These were easy phenotypes to assess, so the venom components that caused the symptomatology were purified by undergraduates at the University of Utah, particularly Craig Clark, who developed the assay, and J. Michael McIntosh, who analyzed C. magus venom and found a similar shaker activity. The amino acid sequences were determined in collaboration with William R. Gray at the University of Utah. The chemical synthesis of these peptides was carried out by Jean Rivier at the Salk Institute.
The two shaker peptides were 25–27 amino acids in length, with three disulfide bonds. They were extremely potent at eliciting the shaker phenotype. We were able to show that these peptides were paralytic to frogs and fish but not to mice. Work by Doju Yoshikami on the neuromuscular junction of the frog demonstrated that the peptides prevented neurotransmitter release; therefore, our hypothesis was that the peptide might be targeting voltage-gated calcium channels at the presynaptic terminus. Preventing calcium entry after an action potential would inhibit neurotransmitter release (21–24). This was demonstrated in a collaboration with Richard W. Tsien’s laboratory (25).
Thus, the first series of venom peptides that were purified and characterized included three from C. geographus, which were paralytic to both fish and frogs, targeted to the postsynaptic nAChR (α-conotoxin GI), to muscle sodium channels (μ-conotoxin GIIIA), and to presynaptic calcium channels that mediated neurotransmitter release (ω-conotoxin GVIA). Ultimately, we showed that toxins in C. magus venom differed significantly in their amino acid sequences from those in the C. geographus homologs but worked by the same mechanisms. At the time, other pharmacological agents targeting nicotinic receptors and sodium channels had been characterized, such as α-bungarotoxin from the Formosan banded krait and tetrodotoxin from the puffer fish, both of which have had a huge impact on molecular neuroscience. However, no ligand was then known for the voltage-gated calcium channels required for neurotransmitter release at the vertebrate neuromuscular junction; the collaboration with the Tsien laboratory showed that ω-conotoxin GVIA was highly selective for only one subclass of mammalian calcium channels, the N-type calcium channel. It was the discovery of ω-conotoxin GVIA that first attracted the attention of the wider neuroscience community to Conus venom peptides, because GVIA quickly became a standard pharmacological reagent for studying synaptic transmission (26). There are now thousands of research papers in the literature on the use of ω-conotoxin GVIA—the impact of this peptide’s wide availability was so pervasive that many molecular neuroscientists use the term conotoxin to refer only to ω-conotoxin GVIA.
The broad use of ω-conotoxin GVIA and its homolog from C. magus venom, ω-conotoxin MVIIA, in the late 1980s ultimately led to a precise definition at both the genetic and the protein levels of the molecular target of ω-conotoxins, the high-voltage-gated calcium channel Cav2.2, or the N-type calcium channel. How ω-conotoxin MVIIA became an approved drug for pain is a story with one major character, George Miljanich, who had been studying the release of the neurotransmitter acetylcholine in the electric organ of the Pacific electric ray, Torpedo californica, at his lab at the University of Southern California. We had sent him ω-conopeptides, and when he transferred to Neurex (a biotech company in Menlo Park, California, that specialized in neuropeptides), George persuaded the management to explore the commercial potential of conopeptides. Because ω-conotoxins were calcium channel inhibitors, they might be neuroprotective. Neurex assessed these peptides in animal models of stroke (the peptides were only modestly effective). While these studies were in progress, the systematic characterization of ω-conotoxins included radiolabeling the peptides and defining the anatomical loci to which the peptides bound. This study revealed that ω-conotoxins bound to a specific region of the spinal cord, the dorsal horn, at the layer where the synaptic connections between pain fibers and spinal cord neurons were located. This result strongly suggested that a key synapse in the pain circuitry might be inhibited by the peptide and that ω-conotoxins might have analgesic potential.
Neurex explored ω-conotoxins as candidate drugs for pain; the initial studies led to a focus on ω-conotoxin MVIIA from C. magus, because it is reversible and therefore less dangerous for a clinician to administer. George Miljanich shepherded the project on a long and convoluted road, and ultimately the peptide was approved by the US Food and Drug Administration (FDA) in 2004 as a drug for intractable pain. The commercial drug, Prialt, is chemically identical to the natural peptide. It is pumped into the intrathecal space and is efficacious in a significant fraction of patients with pain that is unresponsive to opioids.
The Unpredictability of Biology: Recent Surprises
The biology of individual cone snail species has been used to guide new discovery. Uncovering what the snails have evolved constantly surprises us and is what makes integrating different facets of biological experimental science so absorbing. One example is the discovery by Helena Safavi-Hemami that C. geographus, the species we had studied most intensively, releases insulin into the water as it approaches its fish prey (27); this is the smallest functional insulin discovered yet. Thus, to debilitate potential prey, the snail has evolved venom components that target not only the nervous system but also the energy metabolism of fish. The insulin secreted into the water makes fish progressively more hypoglycemic and therefore unable to escape.
Human insulin is designed to persist in the blood to optimize its diverse physiological effects, and consequently, all mammalian insulins are secreted as hexamers that dissociate into the monomers that bind the receptor. The cone snail, however, wants the insulin it secretes into the water to act as quickly as possible; therefore, venom insulin is monomeric so it can act immediately and potently on the fish insulin receptor (28). This fast-acting insulin has led to the design of a monomeric analog of human insulin as a fast-acting drug lead for diabetes (29). Since the discovery of the venom insulin in C. geographus, a diverse set of venom insulins in other cone snail species have been identified (30, 31).
Even more unexpected has been that the work on cone snail venom has potential application for the coronavirus disease 2019 (COVID-19) pandemic. In contrast to previous biomedical applications of conopeptides, the cone snails on which this is based are not fish-hunting but worm- and snail-hunting species. This unanticipated development is based on prior work by Bruce G. Livett and coworkers (32, 33) in Australia—they discovered an α-conopeptide targeted to nicotinic receptors that alleviated neuropathic pain and accelerated the recovery of injured neurons. This peptide, Vc1.1, from the snail-hunting species Conus victoriae, was targeted to the α9α10 nicotinic receptor (34). Another conopeptide, α-conotoxin RgIA, from a worm-hunting cone snail, Conus regius (35), targeted this receptor with even greater potency and specificity and effectively prevented nerve injury and chronic pain after chemotherapy (34, 36, 37). The bioactivity of these peptides appears to occur through the prevention of inflammation resulting from chemotherapy or nerve injury; immune cells are inhibited from detecting extracellular ATP, and as a consequence, this also has potential as a therapeutic in the COVID-19 pandemic. Some of the most life-threatening effects of severe acute respiratory syndrome-related coronavirus-2 (SARS-CoV-2) infection are believed to be due to runaway inflammation. Recently, derivatives of α-conotoxin RgIA were tested on immune cells in the lung and the initial results suggest they could prevent inflammatory pathology, particularly acute respiratory distress syndrome, which has led to many of the human fatalities during the COVID-19 pandemic. Thus, as new peptides are characterized, potential biomedical applications of surprising diversity emerge: diabetes, cancer chemotherapy, and even the COVID-19 pandemic.
AN UNUSUAL NICHE IN PHARMACOLOGY
My research career has led to pharmacology as my core scientific discipline. I remain keenly interested in the clinical applications of pharmacology, and I am delighted that cone snail venom peptides continue to exhibit strong potential as drug leads. Nevertheless, my focus is on advancing pharmacology as a basic science in order to accelerate our understanding of the biological function at multiple levels. The dominant present intellectual approach is genetic: The use of model organisms and advances in the sequencing and manipulation of DNA have led to the rapid development of genetic tools to investigate function. Insights into function are clearly obtained by manipulating genes, but our understanding of the biological function would proceed much more rapidly if the genetic approach were coupled with robust pharmacology. This is particularly true of efforts to understand the nervous system and how normal function goes awry in the pathologies of the nervous system that are some of the most intractable health problems of our time. To address function, we need a pharmacological arsenal that is as sophisticated as the technology available for genetics and genomics.
In our efforts to understand cone snail venom peptides and their molecular targets (generally receptors and ion channels), a core conceptual issue emerged that is seldom addressed. Many signal transduction proteins, particularly ion channels, comprise multiple subunits, with each subunit being the product of an individual gene. Thus, a functional voltage-gated potassium channel is a tetramer, which means that a fully assembled functional channel might be the homomeric product of one gene, a heteromer of two different gene products, and, in principle, a combination of the products of as many as four different genes. Such combinatorial possibilities generate an enormous potential for complexity that cannot be easily addressed through genetics alone. Mutation of one gene encoding a potassium channel subunit could affect several different functional tetrameric channels that are molecularly (and presumably functionally) distinct from each other, leading to a quagmire in interpreting the mutant phenotypes obtained. To elucidate this complexity, complementary pharmacology must be able to systematically define individual functional heteromeric channels.
Recent work using one highly selective conopeptide (38) has initially defined the impressive scale of the problem of elucidating the functional roles of each molecular isoform of a large ion channel family (such as voltage-gated potassium channels) in different cell types. This study, carried out in collaboration with Heinrich Terlau at the University of Kiel, used a Conus venom peptide, κMRIIIJ, that selectively targeted a specific heteromeric voltage-gated potassium channel isoform. This study demonstrated that different specific heteromeric combinations are present in the divergent subtypes of neurons present in the same tissue. Using a combination of calcium imaging, pharmacology, and single-cell transcriptomes, researchers can assess the function of specific ion channel isoforms in individual types of sensory neurons (L.S. Leavitt, K. Tsourmas, R. Alluri, M.J. Giacobassi, V.H. Rockhill, et al., manuscript submitted); such a functional assessment would not have been possible through genetics alone.
The ability to simultaneously analyze multiple cell types in a tissue through calcium imaging, using highly specific pharmacological agents (such as Conus venom peptides), combined with transcriptomics and proteomics, an integrated discipline we call pharmaconomics, makes it possible to define in unprecedented detail how and when molecular changes occur as a pathology develops. Being able to chart cell-specific molecular alterations in a tissue relevant to a pathology (39) as the pathology develops would make feasible a systematic query of whether the particular molecular change observed is causative for the further progression to the pathological state. The availability of highly specific pharmacological agents to counteract any uncovered causative molecular changes would provide a foundation for identifying whole new classes of molecular targets and, through the development of highly selective ligands for each of these targets, could lead to novel drugs. Such drugs would be the basis for preventative therapeutics, preventing the disease state instead of attempting to alleviate symptoms after a pathological state becomes established.
This ambitious agenda, which involves the application of conopeptides, has been the major focus of our highly collaborative research program. As I finish this review (May 2020), the world has suddenly changed and so our priorities must necessarily adapt. We are intensively exploring how to use the pharmaconomics approach described above to understand what happens to lung tissue that leads to the need for ventilators after infection by a respiratory tract virus. The world will need preventative therapeutics, so that in future pandemics, as the first mild symptoms appear, the complete collapse of respiratory function might be alleviated or even prevented.

CONCLUDING THOUGHTS
At the start of my scientific career, the word pharmacology would conjure for me, like most people, a picture of bottles of pills in a drugstore. This biographical history documents my complicated journey to the very different perception that I hold today. When I think about pharmacology, what comes to mind is the essential core of science that will be increasingly indispensable for seamlessly integrating multiple levels of biological function, from molecules to systems to the whole organism.
This story has been reproduced in full from the Annual Review of Pharmacology and Toxicology, 2021.
