
DNA metabarcoding is the large-scale taxonomic identification of complex environmental samples via analysis of DNA sequences for short regions of one or a few genes.
The technique is widely used to determine wild animal diets, but whether this technique provides accurate, quantitative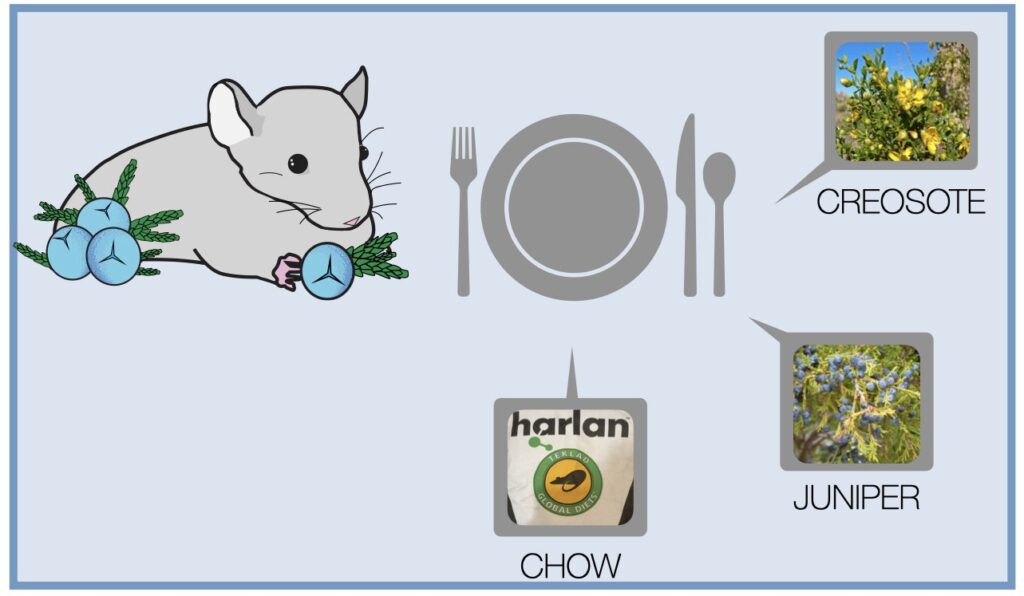 measurements is still under debate.
measurements is still under debate.
A team of SBS researchers set out to test researchers’ ability to accurately estimate the abundance of dietary items using metabarcoding by feeding wild-caught desert woodrats (Neotoma lepida) a controlled diet. Then, using metabarcoding, they compared the representation of items in the original diet samples to that in samples of feces. This was done to test the sensitivity and accuracy of diet metabarcoding and also the performance of different bioinformatic pipelines and the researcher’s ability to correct sequence counts.
The study was recently published in Molecular Ecology Resources with lead author Tess Stapleton (Dearing lab) who successfully defended her PhD thesis recently.
Her results revealed the successes and limitations of this technique in plant-eating animals with a single-compartment stomach, also known as “non-ruminant herbivores.”
While metabarcoding successfully detected rare diet components, researchers found it was contingent upon whether primers could amplify these taxa. Scientists know that one benefit of the technique compared to more traditional dietary analysis is its ability to detect and accurately assign taxonomy to rare species. Such was the case with detecting toxic creosote at proportions as low as 1%, but in the case of cactus–even when it was the only diet component–the technique failed to detect it.

This is due, the researchers conjecture, to what’s known as a “primer mismatch,” which brings into question metabarcoding primers commonly referred to as “universal” due to their ability to amplify DNA from many different species. However, the team determined that these universal primers do not match all species and can completely miss important components of an animal’s diet.
“Although we were able to amplify cactus DNA when it was the only component of the diet,” the paper’s discussion reads, “it was lost once more than one plant was present in the mixture.”
These complications to the technique of metabarcoding need to be navigated before accurate detections can occur, especially in wild, mammalian herbivores who typically do not feed on a single plant.
Critically, they report, “while correction factors can account for errors due to differential amplification, error due to differential digestion and other biological factors will be more difficult to correct.” And yet, “[c]ollectively, our results have important implications for the use of metabarcoding in herbivore diet determination.”
As far as implications of the research, Stapleton says, “We were surprised to see that not only could we detect plants when they were a very rare component of the diet, but we could also statistically differentiate between plants that made up a low proportion of the diet and a high proportion of the diet, which means that this technique can be used in a quantitative way.”
You can read the entire article, from which this summary was derived, including the detailed discussion here.
In addition to Stapleton, other authors of the study include Sara Weinstein, Robert Greenhalgh, and M. Denise Dearing.
